pacman::p_load(tmap, sf, sfdep, tidyverse, knitr)In-class Exercise 2: Spatial
GLSA
Getting Started - Import packages
This function calls pacman to load sf, tidyverse, tmap, knitr packages;
tmap: For thematic mapping; powerful mapping packagesf: for geospatial data handling, but also geoprocessing: buffer, point-in-polygon count, etc- batch processing over GIS packages; can handle tibble format
sfdep: creates space-time cube, EHSA; replaces spdeptidyverse: for non-spatial data handling; commonly used R packageknitr: generates html table
Loading the data
Hunan: geospatial dataset in ESRI shapefile format- use of
st_read()to import assfdata.frame$geometrycolumn is actually a list inside thedfcell; that’s the power of the tibble dataframe- “features” of
simple featuresrefers to geometric features eg point line curve etc
- note projection is
WGS84; see `88
- use of
hunan2012: attribute format in csv format- use of
read_csv()astbl_dfdata.frame
- use of
- !IMPORTANT! to retain geometry, you must left join to the
sfdataframe (eg you can also hunan2012 right join hunan)- without
sfdataframe, normal tibble dataframe will drop the geometry column
- without
show code
hunan <- st_read(dsn = "data/geospatial",
layer = "Hunan")Reading layer `Hunan' from data source
`C:\1darren\ISSS624\In-class_Ex\In-class_Ex2\data\geospatial'
using driver `ESRI Shapefile'
Simple feature collection with 88 features and 7 fields
Geometry type: POLYGON
Dimension: XY
Bounding box: xmin: 108.7831 ymin: 24.6342 xmax: 114.2544 ymax: 30.12812
Geodetic CRS: WGS 84show code
hunan2012 <- read_csv("data/aspatial/Hunan_2012.csv")
hunan_GDPPC <- left_join(hunan,hunan2012)%>%
select(1:4, 7, 15)Plot a chloropleth of GDPPC
show code
#qtm(hunan, "GDPPC") +
# tm_layout(main.title = "GDPPC", main.title.position = "right")
tm_shape(hunan_GDPPC) +
tm_fill(col = "GDPPC",
style = "pretty",
palette = "Blues",
title = "GDPPC") +
tm_borders(alpha = 0.5) +
tm_layout(main.title = "GDPPC",
inner.margins = c(0.1, 0.1, 0.1, 0.1),
outer.margins = c(0.1, 0.1, 0.1, 0.1)
) +
tm_grid(alpha = ) 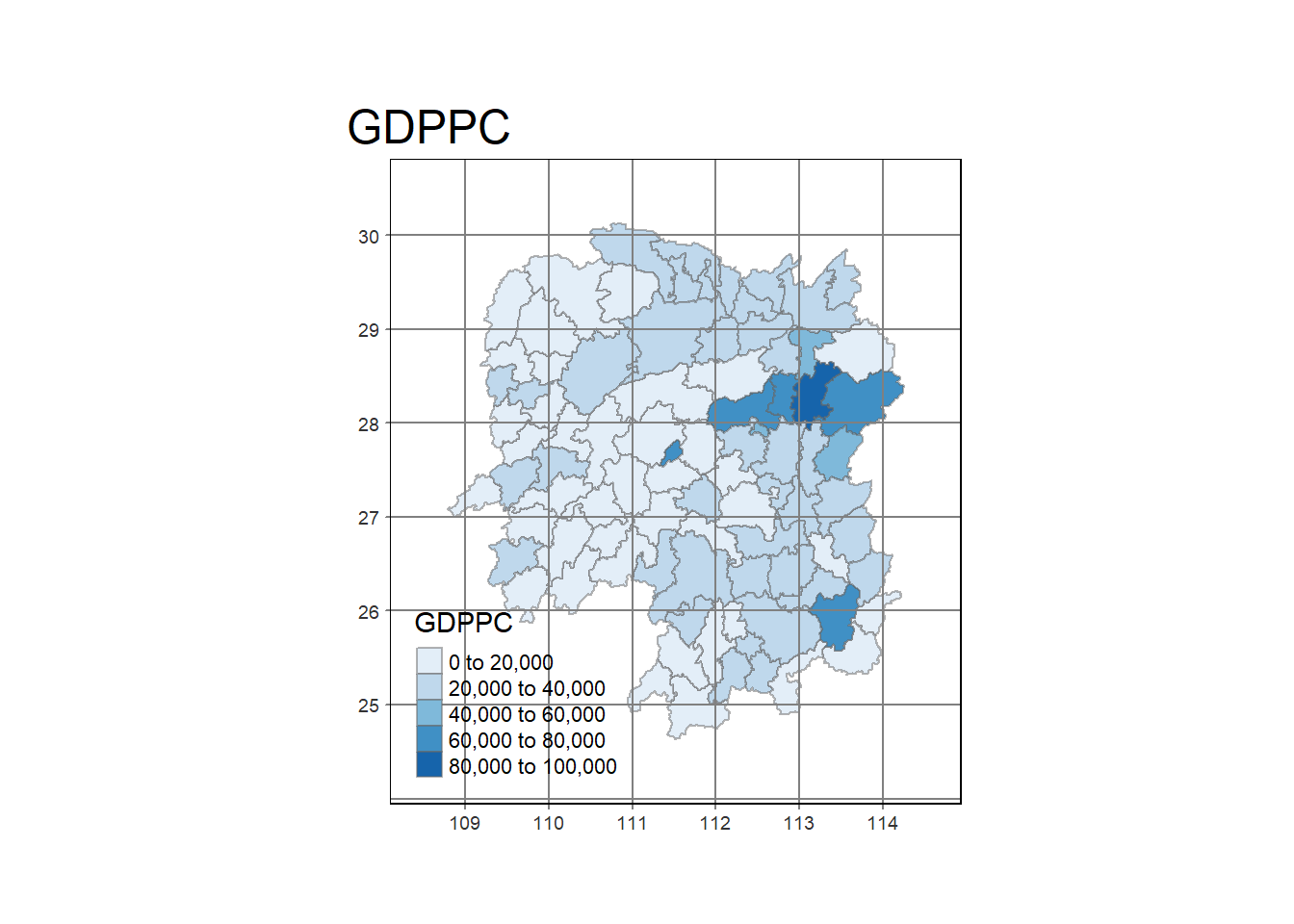
Deriving QUEEN contiguity weights
mutateis function that creates new column from previous column datasst_contiguitycreatesnbneighbour matrix (QUEEN contiguity, by default)st_weightscreates row-standardised weights (style="W") fromnbobject- One-step function using
sfdep; a wrapper forspdepbut writes output intosfdataframe
show code
wm_q <- hunan_GDPPC %>%
mutate(nb = st_contiguity(geometry),
wt = st_weights(nb, style = "W"),
.before = 1)Computing Global Moran’s I
- below is “old_style”
show code
# moran_i = global_moran(
# hunan_GDPPC$GDPPC,
# hunan_GDPPC$nb,
# hunan_GDPPC$wt
# )
# glimpse(moran_i)Computing Local Moran’s I
- Monte Carlo: simulation more accurate, calculate Local Moran’s I using
unnestis needed to turn the output oflocal_moraninto individual columns inlisalocal_moranwill create a group table; a separate list of columns that’s hard to read
- several high/low options for Moran’s I
$meanis default;$mediancan be used if distribution is highly skewed (eg skew high == biased to right)
show code
#
# lisa <- wm_q %>%
# mutate(localmoran = local_moran(GDPPC, nb, wt, nsim=99),
# .before = 1) %>%
# unnest(localmoran)
# glimpse(lisa)