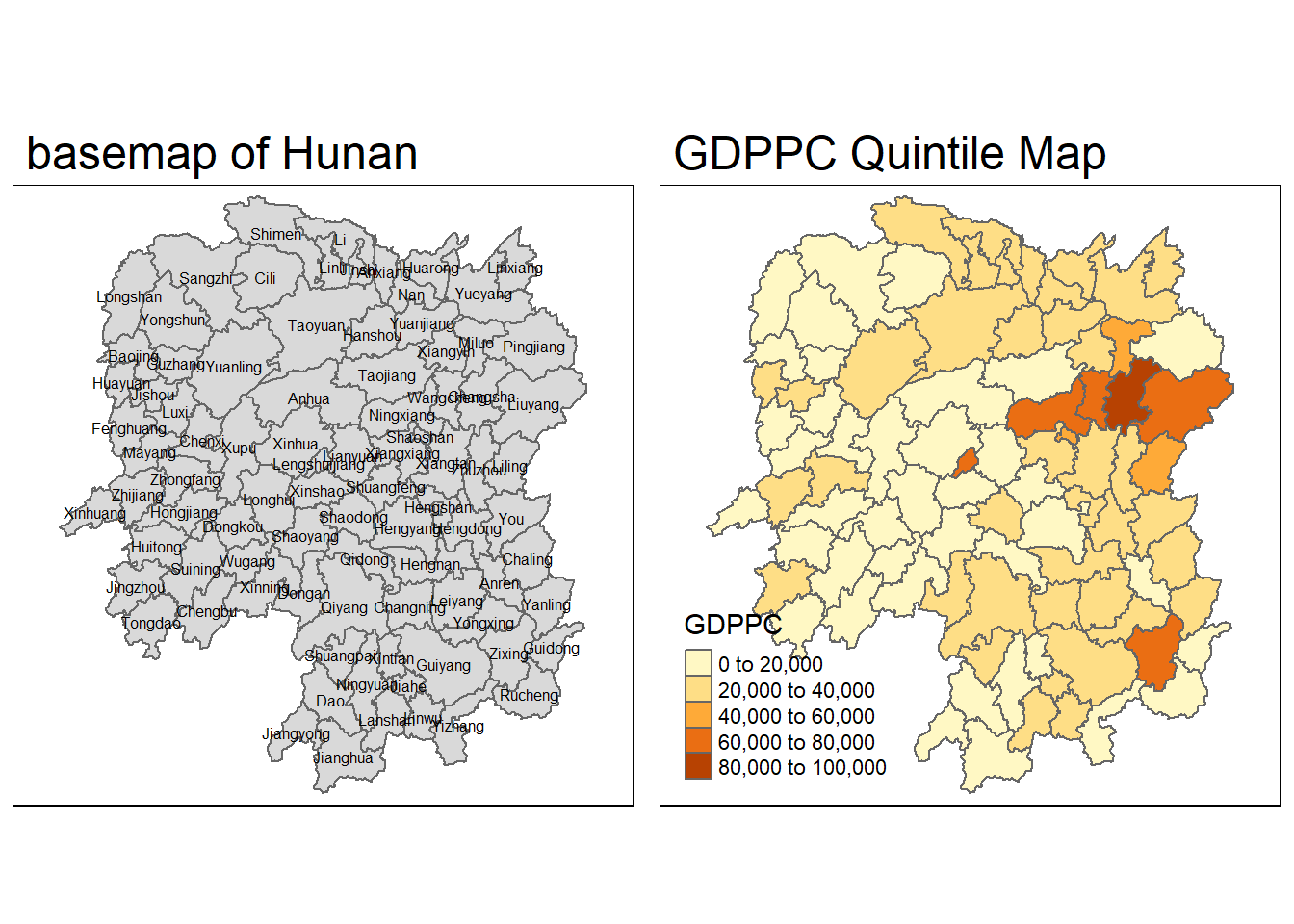
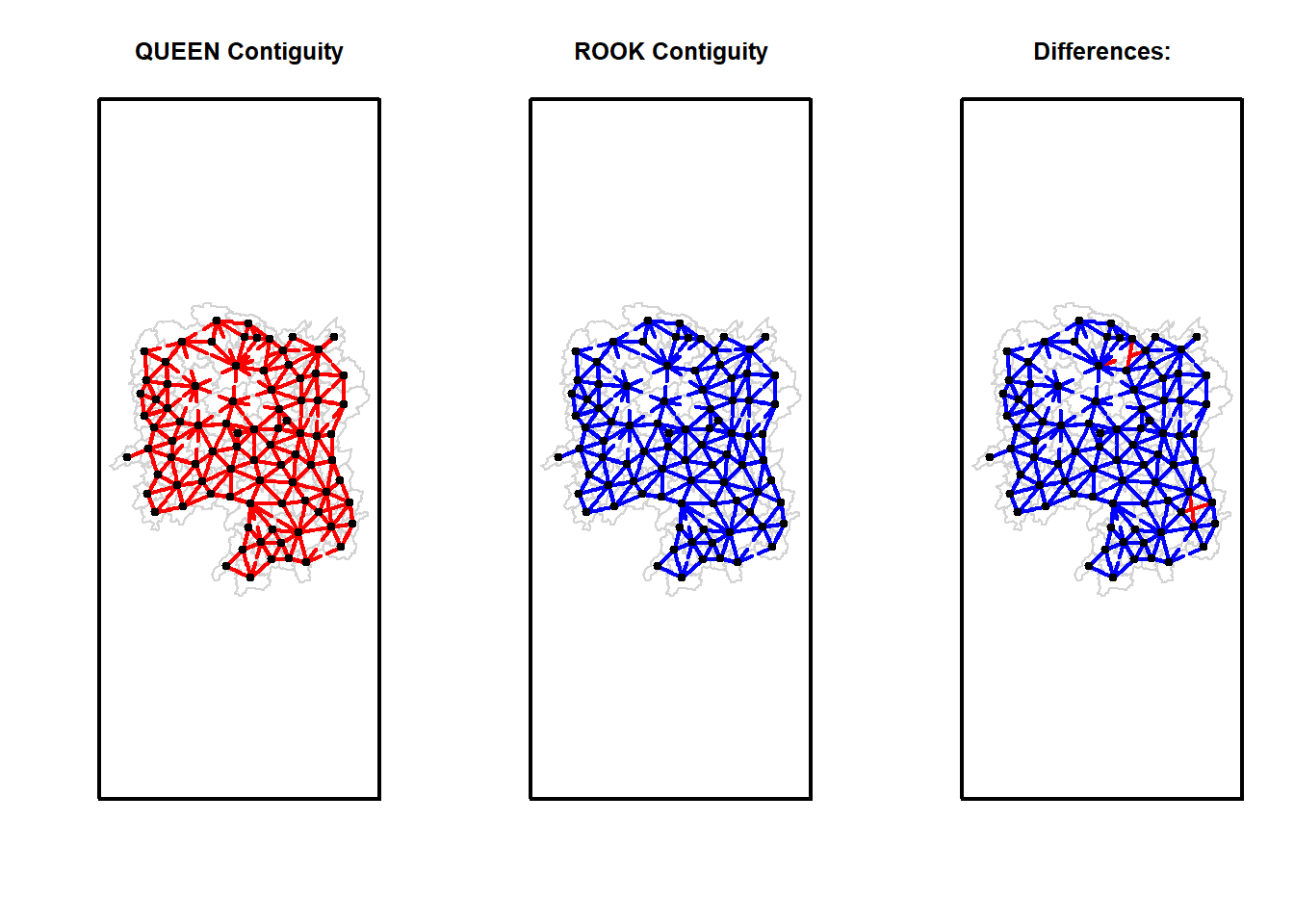
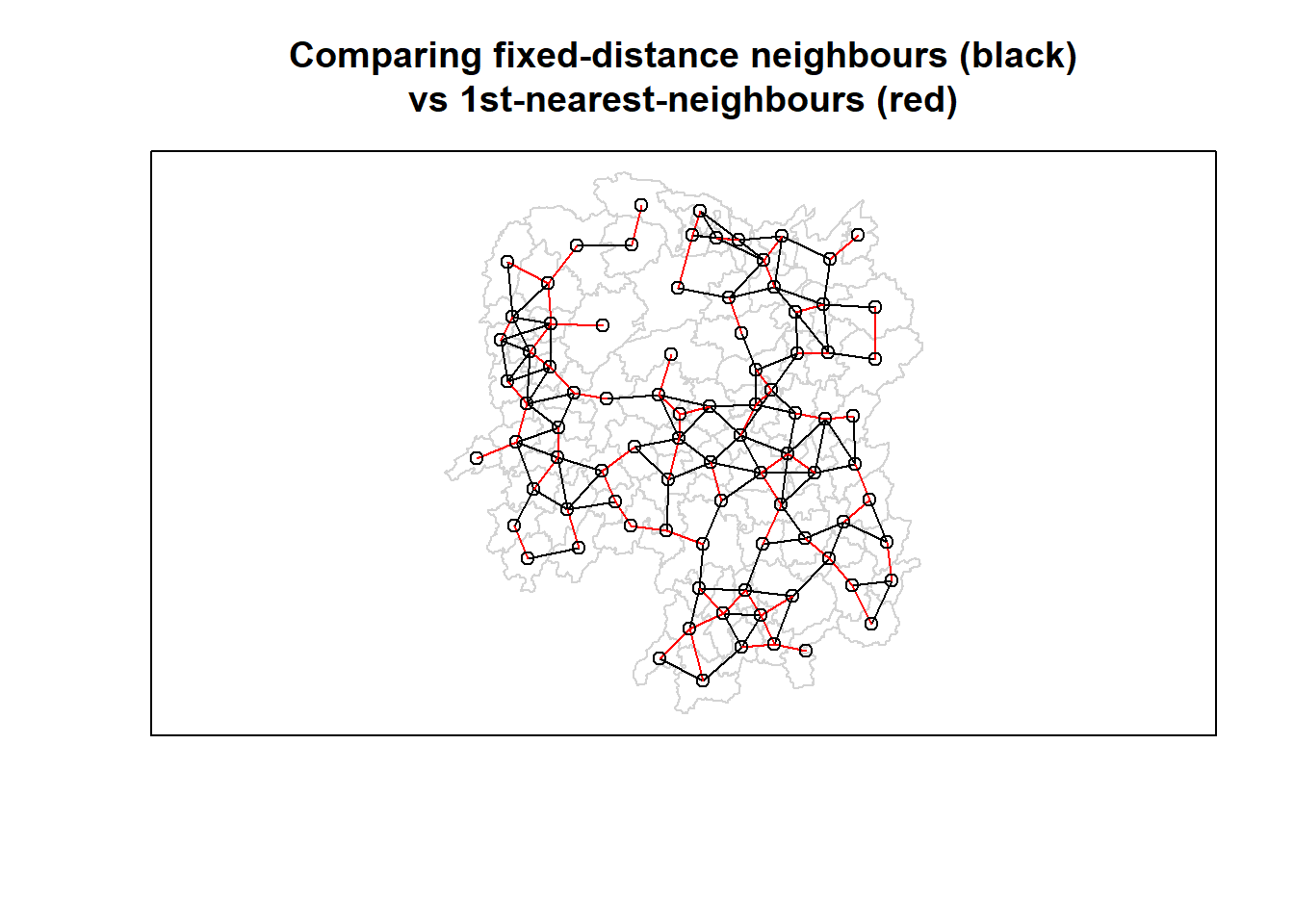
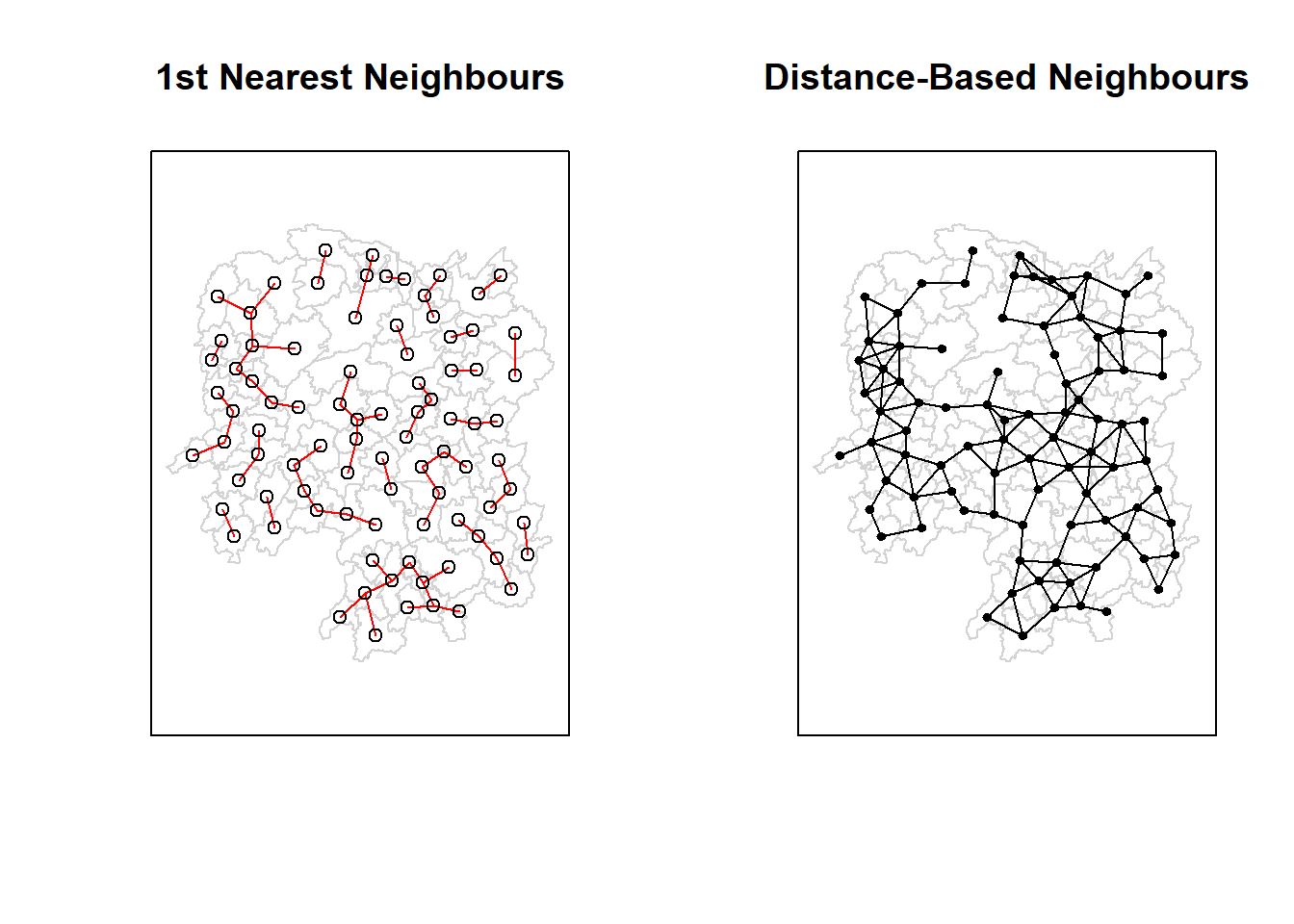
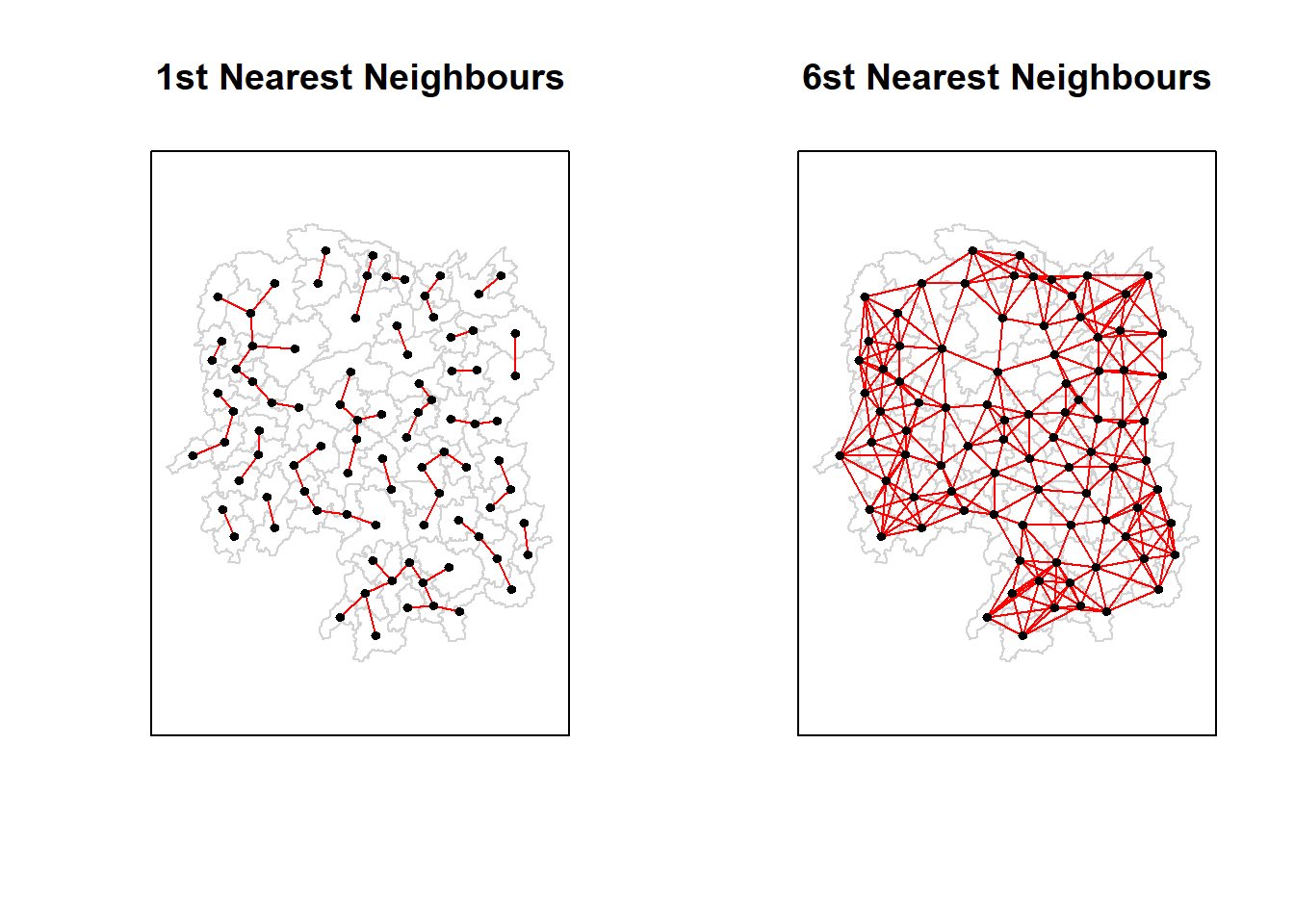
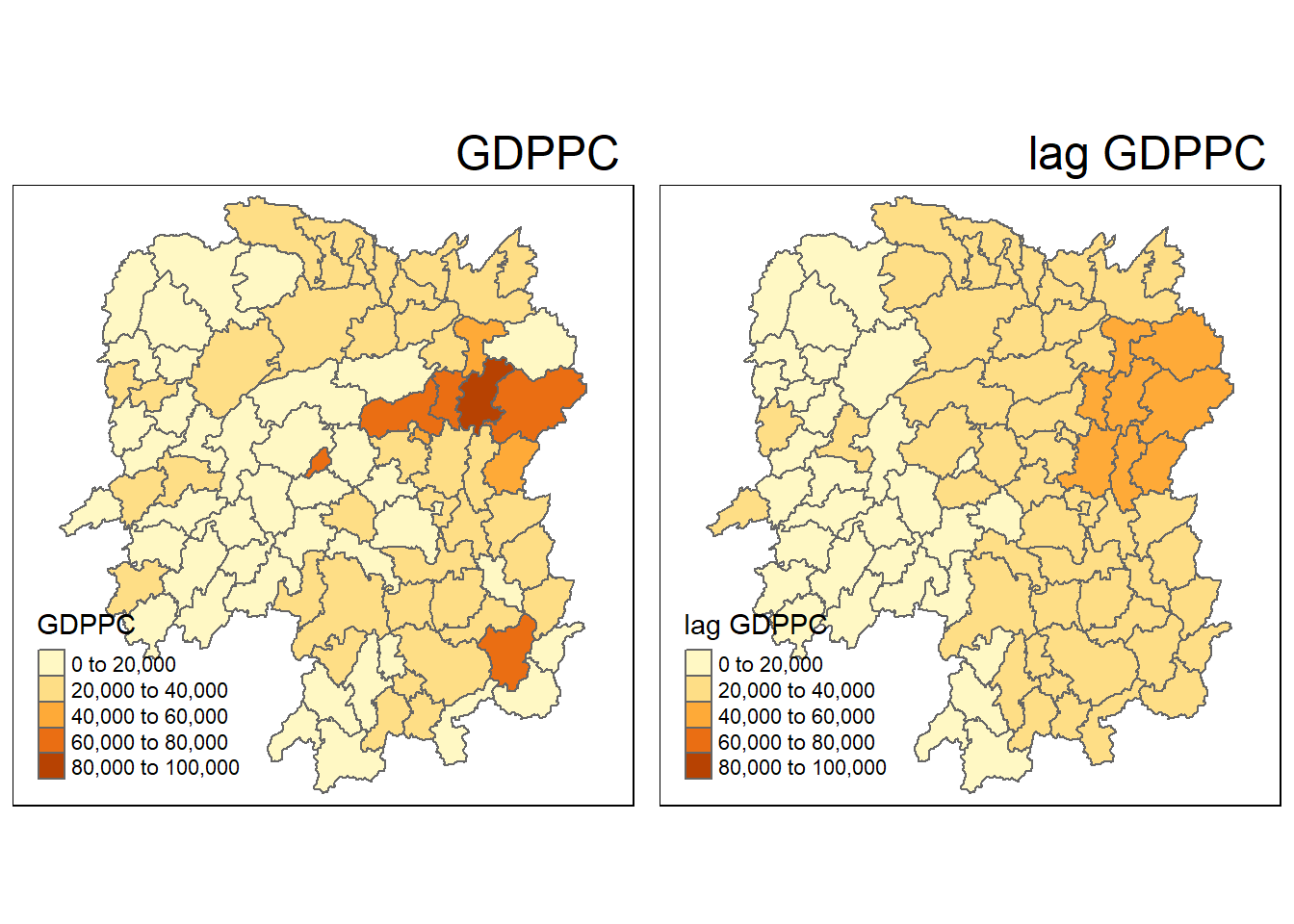
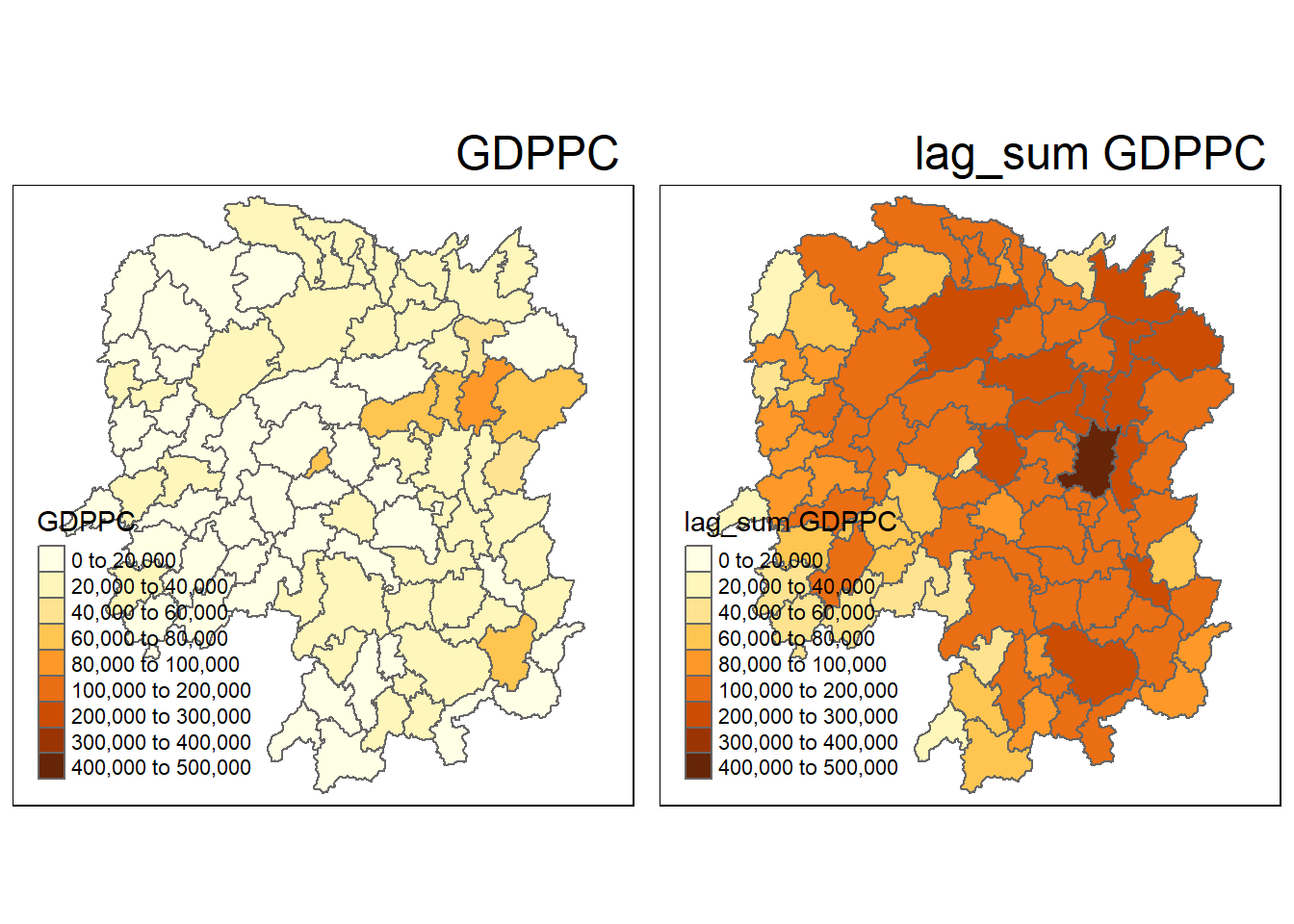
| Anxiang |
24847.20 |
124236 |
24650.50 |
POLYGON ((112.0625 29.75523… |
| Hanshou |
22724.80 |
113624 |
22434.17 |
POLYGON ((112.2288 29.11684… |
| Jinshi |
24143.25 |
96573 |
26233.00 |
POLYGON ((111.8927 29.6013,… |
| Li |
27737.50 |
110950 |
27084.60 |
POLYGON ((111.3731 29.94649… |
| Linli |
27270.25 |
109081 |
26927.00 |
POLYGON ((111.6324 29.76288… |
| Shimen |
21248.80 |
106244 |
22230.17 |
POLYGON ((110.8825 30.11675… |
| Liuyang |
43747.00 |
174988 |
47621.20 |
POLYGON ((113.9905 28.5682,… |
| Ningxiang |
33582.71 |
235079 |
37160.12 |
POLYGON ((112.7181 28.38299… |
| Wangcheng |
45651.17 |
273907 |
49224.71 |
POLYGON ((112.7914 28.52688… |
| Anren |
32027.62 |
256221 |
29886.89 |
POLYGON ((113.1757 26.82734… |
| Guidong |
32671.00 |
98013 |
26627.50 |
POLYGON ((114.1799 26.20117… |
| Jiahe |
20810.00 |
104050 |
22690.17 |
POLYGON ((112.4425 25.74358… |
| Linwu |
25711.50 |
102846 |
25366.40 |
POLYGON ((112.5914 25.55143… |
| Rucheng |
30672.33 |
92017 |
25825.75 |
POLYGON ((113.6759 25.87578… |
| Yizhang |
33457.75 |
133831 |
30329.00 |
POLYGON ((113.2621 25.68394… |
| Yongxing |
31689.20 |
158446 |
32682.83 |
POLYGON ((113.3169 26.41843… |
| Zixing |
20269.00 |
141883 |
25948.62 |
POLYGON ((113.7311 26.16259… |
| Changning |
23901.60 |
119508 |
23987.67 |
POLYGON ((112.6144 26.60198… |
| Hengdong |
25126.17 |
150757 |
25463.14 |
POLYGON ((113.1056 27.21007… |
| Hengnan |
21903.43 |
153324 |
21904.38 |
POLYGON ((112.7599 26.98149… |
| Hengshan |
22718.60 |
113593 |
23127.50 |
POLYGON ((112.607 27.4689, … |
| Leiyang |
25918.80 |
129594 |
25949.83 |
POLYGON ((112.9996 26.69276… |
| Qidong |
20307.00 |
142149 |
20018.75 |
POLYGON ((111.7818 27.0383,… |
| Chenxi |
20023.80 |
100119 |
19524.17 |
POLYGON ((110.2624 28.21778… |
| Zhongfang |
16576.80 |
82884 |
18955.00 |
POLYGON ((109.9431 27.72858… |
| Huitong |
18667.00 |
74668 |
17800.40 |
POLYGON ((109.9419 27.10512… |
| Jingzhou |
14394.67 |
43184 |
15883.00 |
POLYGON ((109.8186 26.75842… |
| Mayang |
19848.80 |
99244 |
18831.33 |
POLYGON ((109.795 27.98008,… |
| Tongdao |
15516.33 |
46549 |
14832.50 |
POLYGON ((109.9294 26.46561… |
| Xinhuang |
20518.00 |
20518 |
17965.00 |
POLYGON ((109.227 27.43733,… |
| Xupu |
17572.00 |
140576 |
17159.89 |
POLYGON ((110.7189 28.30485… |
| Yuanling |
15200.12 |
121601 |
16199.44 |
POLYGON ((110.9652 28.99895… |
| Zhijiang |
18413.80 |
92069 |
18764.50 |
POLYGON ((109.8818 27.60661… |
| Lengshuijiang |
14419.33 |
43258 |
26878.75 |
POLYGON ((111.5307 27.81472… |
| Shuangfeng |
24094.50 |
144567 |
23188.86 |
POLYGON ((112.263 27.70421,… |
| Xinhua |
22019.83 |
132119 |
20788.14 |
POLYGON ((111.3345 28.19642… |
| Chengbu |
12923.50 |
51694 |
12365.20 |
POLYGON ((110.4455 26.69317… |
| Dongan |
14756.00 |
59024 |
15985.00 |
POLYGON ((111.4531 26.86812… |
| Dongkou |
13869.80 |
69349 |
13764.83 |
POLYGON ((110.6622 27.37305… |
| Longhui |
12296.67 |
73780 |
11907.43 |
POLYGON ((110.985 27.65983,… |
| Shaodong |
15775.17 |
94651 |
17128.14 |
POLYGON ((111.9054 27.40254… |
| Suining |
14382.86 |
100680 |
14593.62 |
POLYGON ((110.389 27.10006,… |
| Wugang |
11566.33 |
69398 |
11644.29 |
POLYGON ((110.9878 27.03345… |
| Xinning |
13199.50 |
52798 |
12706.00 |
POLYGON ((111.0736 26.84627… |
| Xinshao |
23412.00 |
140472 |
21712.29 |
POLYGON ((111.6013 27.58275… |
| Shaoshan |
39541.00 |
118623 |
43548.25 |
POLYGON ((112.5391 27.97742… |
| Xiangxiang |
36186.60 |
180933 |
35049.00 |
POLYGON ((112.4549 28.05783… |
| Baojing |
16559.60 |
82798 |
16226.83 |
POLYGON ((109.7015 28.82844… |
| Fenghuang |
20772.50 |
83090 |
19294.40 |
POLYGON ((109.5239 28.19206… |
| Guzhang |
19471.20 |
97356 |
18156.00 |
POLYGON ((109.8968 28.74034… |
| Huayuan |
19827.33 |
59482 |
19954.75 |
POLYGON ((109.5647 28.61712… |
| Jishou |
15466.80 |
77334 |
18145.17 |
POLYGON ((109.8375 28.4696,… |
| Longshan |
12925.67 |
38777 |
12132.75 |
POLYGON ((109.6337 29.62521… |
| Luxi |
18577.17 |
111463 |
18419.29 |
POLYGON ((110.1067 28.41835… |
| Yongshun |
14943.00 |
74715 |
14050.83 |
POLYGON ((110.0003 29.29499… |
| Anhua |
24913.00 |
174391 |
23619.75 |
POLYGON ((111.6034 28.63716… |
| Nan |
25093.00 |
150558 |
24552.71 |
POLYGON ((112.3232 29.46074… |
| Yuanjiang |
24428.80 |
122144 |
24733.67 |
POLYGON ((112.4391 29.1791,… |
| Jianghua |
17003.00 |
68012 |
16762.60 |
POLYGON ((111.6461 25.29661… |
| Lanshan |
21143.75 |
84575 |
20932.60 |
POLYGON ((112.2286 25.61123… |
| Ningyuan |
20435.00 |
143045 |
19467.75 |
POLYGON ((112.0715 26.09892… |
| Shuangpai |
17131.33 |
51394 |
18334.00 |
POLYGON ((111.8864 26.11957… |
| Xintian |
24569.75 |
98279 |
22541.00 |
POLYGON ((112.2578 26.0796,… |
| Huarong |
23835.50 |
47671 |
26028.00 |
POLYGON ((112.9242 29.69134… |
| Linxiang |
26360.00 |
26360 |
29128.50 |
POLYGON ((113.5502 29.67418… |
| Miluo |
47383.40 |
236917 |
46569.00 |
POLYGON ((112.9902 29.02139… |
| Pingjiang |
55157.75 |
220631 |
47576.60 |
POLYGON ((113.8436 29.06152… |
| Xiangyin |
37058.00 |
185290 |
36545.50 |
POLYGON ((112.9173 28.98264… |
| Cili |
21546.67 |
64640 |
20838.50 |
POLYGON ((110.8822 29.69017… |
| Chaling |
23348.67 |
70046 |
22531.00 |
POLYGON ((113.7666 27.10573… |
| Liling |
42323.67 |
126971 |
42115.50 |
POLYGON ((113.5673 27.94346… |
| Yanling |
28938.60 |
144693 |
27619.00 |
POLYGON ((113.9292 26.6154,… |
| You |
25880.80 |
129404 |
27611.33 |
POLYGON ((113.5879 27.41324… |
| Zhuzhou |
47345.67 |
284074 |
44523.29 |
POLYGON ((113.2493 28.02411… |
| Sangzhi |
18711.33 |
112268 |
18127.43 |
POLYGON ((110.556 29.40543,… |
| Yueyang |
29087.29 |
203611 |
28746.38 |
POLYGON ((113.343 29.61064,… |
| Qiyang |
20748.29 |
145238 |
20734.50 |
POLYGON ((111.5563 26.81318… |
| Taojiang |
35933.71 |
251536 |
33880.62 |
POLYGON ((112.0508 28.67265… |
| Shaoyang |
15439.71 |
108078 |
14716.38 |
POLYGON ((111.5013 27.30207… |
| Lianyuan |
29787.50 |
238300 |
28516.22 |
POLYGON ((111.6789 28.02946… |
| Hongjiang |
18145.00 |
108870 |
18086.14 |
POLYGON ((110.1441 27.47513… |
| Hengyang |
21617.00 |
108085 |
21244.50 |
POLYGON ((112.7144 26.98613… |
| Guiyang |
29203.89 |
262835 |
29568.80 |
POLYGON ((113.0811 26.04963… |
| Changsha |
41363.67 |
248182 |
48119.71 |
POLYGON ((112.9421 28.03722… |
| Taoyuan |
22259.09 |
244850 |
22310.75 |
POLYGON ((112.0612 29.32855… |
| Xiangtan |
44939.56 |
404456 |
43151.60 |
POLYGON ((113.0426 27.8942,… |
| Dao |
16902.00 |
67608 |
17133.40 |
POLYGON ((111.498 25.81679,… |
| Jiangyong |
16930.00 |
33860 |
17009.33 |
POLYGON ((111.3659 25.39472… |